STAGBEETLE
SUPLEMENTARY INFORMATION
OTHER INFORMATION
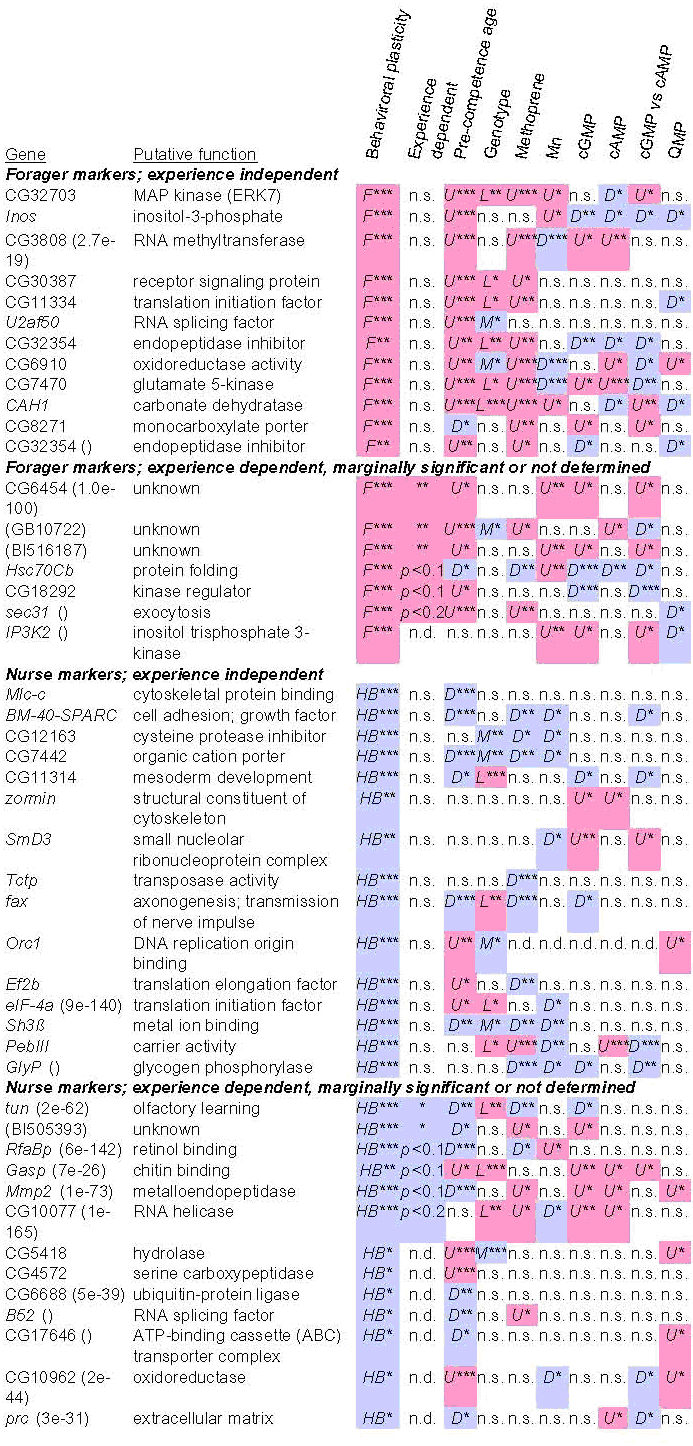
Fig. 4. Candidate
genes for honey bee behavioral plasticity. Shown are a subset of the 100
behavioral marker genes for which we replicated the findings in ref. (1) in the
current study (hive-to-forager differences, p
< 0.05) and have functional (Gene Ontology) annotation. Genes listed without parentheses are putative
Drosophila orthologs
based on reciprocal best blast match. Genes followed by parentheses are best Drosophila match (BLAST e-value
indicated). Two genes listed in parentheses are a predicted gene (GB10722) and
an EST (BI516187) from A. mellifera with no Drosophila
matches at BLAST e-value < 10-5. Color and letter indicates
direction of regulation: up-regulated (U); down-regulated (D); higher in
forager (F), hive bee (HB), ligustica (L), mellifera (M). *, p
< 0.05; **, p < 0.001; ***, p < 1 × 10-6; n.s., not significant; marginally significant p-values are indicated for
experience-dependence. Statistical tests
from Table 1. A total of 16 out of the 100 genes listed here met three
predictions (see text) for genes that could play causal roles in the hive bee
to forager transition.